The binary vectors contain Cas9 (nuclease or nickase) and the gRNA. Cas9 is codon optimized for high expression in monocots and dicots
RNA is driven by the U6 promoter from Arabidopsis thaliana (for dicots) or Oryza sativa (for monocots).
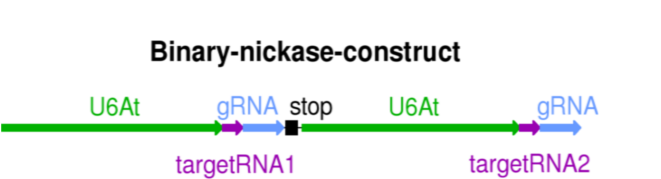
Insertion of the target RNA in the Cas9 nickase vector can be easily done as DNA fragment containing both target RNAs at the ends. The inserted DNA fragment contains the U6 promoter.
Genes used in plant molecular biology sometimes have low expression in plants due to the fact that they have been originally isolated from bacteria which use a different codon usage.
The betaglucuronidase (GUS) gene from E. coli was codon optimized for better expression in monocots and dicots.
To prevent expression from bacteria the StLS1 intron was introduced. The synthetic GUS gene is regulated by the Ubi promoter from maize and introduced into maize embryos by particle gun bombardment. For comparison the GUS gene from Jefferson et al. (EMBO J. 87 Dec) was used, which is 100% identical in amino acid sequence and in the sourrounding of the genes.



| Nr. | Euro | |
|---|---|---|
| 2600 | Cloning of 1 target site into a Cas9 vector | 350 |
| 2601 | Cloning of a second target site into a Cas9 vector | 300 |
| 2602 | Cloning of a third target site into a Cas9 vector | 275 |
| 2610 | Cloning of 1 double target site into a Cas9 vector | 400 |
| 2611 | Cloning of 2 double target sites into a Cas9 vector | 350 |
| 2612 | Cloning of 4 double target sites into a Cas9 vector | 325 |
| 2613 | Cloning of 8 double target sites into a Cas9 vector | 300 |
| 2700 | Cloning of 1 fourfold target sites into a Cas9 vector | 1.200 |
| 2800 | Cloning of 1 sixfold target sites into a Cas9 vector | 2.000 |